Deep learning-based virtual staining of tissue facilitates rapid assessment of breast cancer biomarker
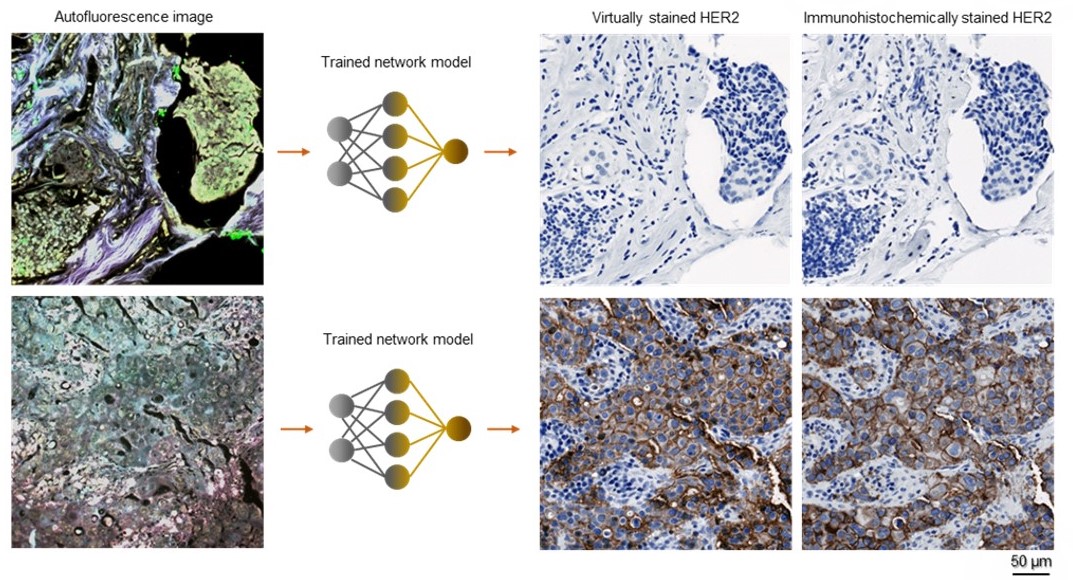
Breast cancer is one the leading causes of cancer death among women globally. Upon breast cancer diagnosis, the testing of HER2 ? a protein that promotes cancer cell growth, is routinely carried out to help assess the cancer prognosis and make HER2-directed treatment plans. A standard HER2 test procedure includes taking the breast biopsy, preparing the tissue specimen into thin microscopic slides, staining/dying the slides with specific chemical reagents that highlight the HER2 proteins, and inspecting the stained slides under an optical microscope to provide the pathological report. However, this standard HER2 staining procedure suffers from high costs and long turn-around time as the staining process requires laborious sample treatment steps (typically ~24 hours) performed by experts in a dedicated laboratory facility.
In a recent work published in BME Frontiers, a UCLA research team developed a computational staining approach powered by deep learning, which performs the HER2 staining without requiring any chemicals. The research team captured the autofluorescence information of the unstained breast tissue, which is naturally emitted by biological structures when they absorb light. They further trained a deep neural network that rapidly transforms these stain-free autofluorescence images into virtual histological images, revealing the accurate color and contrast as if the tissue sections were chemically stained for HER2. This computational staining process takes only a few minutes per sample and does not need expensive facilities or toxic chemicals. Using only a computer, the HER2 staining could be accomplished much faster and cost-effectively, accelerating breast cancer assessments and treatment.
Board-certified pathologists blindly validated this AI-based virtual HER2 staining technique in terms of both its diagnostic value and stain quality. The pathologists confirmed that the deep learning-generated images provide the equivalent diagnostic accuracy for HER2 assessment and have a staining quality comparable to the standard images chemically stained in the laboratory. This deep learning-powered virtual HER2 staining approach eliminates the need for costly, laborious, and time-consuming HER2 staining procedures performed by histology experts and could be extended to staining of other cancer-related biomarkers to accelerate the traditional histopathology and diagnostic workflow in clinical settings.
This research was led by Dr. Aydogan Ozcan, Chancellor’s Professor and Volgenau Chair for Engineering Innovation at UCLA Electrical and Computer Engineering and Bioengineering. The UCLA team collaborated with Dr. Morgan Angus Darrow, Dr. Elham Kamangar, and Dr. Han Sung Lee, breast pathologists at the Department of Pathology and Laboratory Medicine from UC Davis. The other authors of this work include Bijie Bai, Hongda Wang, Yuzhu Li, Kevin de Haan, Francesco Colonnese, Yujie Wan, Jingyi Zuo, Ngan B. Doan, Xiaoran Zhang, Yijie Zhang, Jingxi Li, Xilin Yang, Wenjie Dong and Dr. Yair Rivenson, all affiliated with UCLA.
The National Science Foundation’s Biophotonics Program and NIH/National Center supported the research.
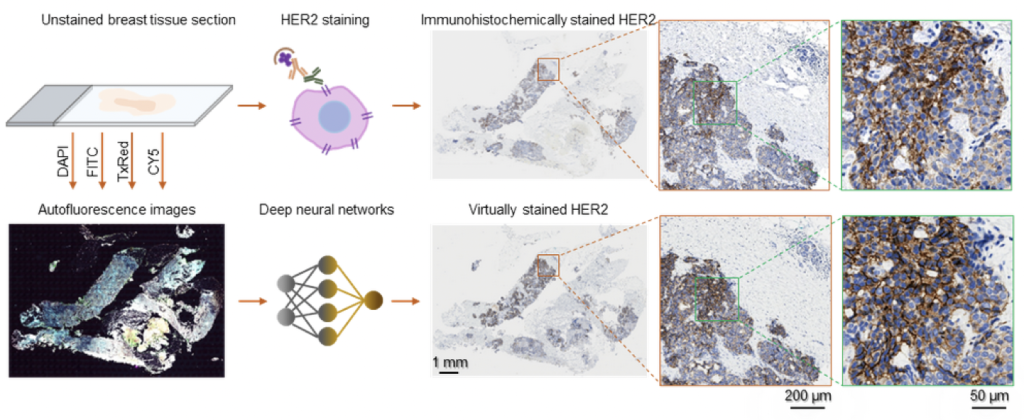
Figure | Virtual HER2 staining of unlabeled breast tissue sections using deep learning. Image credit: Ozcan Lab @ UCLA.
See the article:
Bijie Bai, Hongda Wang, Yuzhu Li, Kevin de Haan, Francesco Colonnese, Yujie Wan, Jingyi Zuo, Ngan B. Doan, Xiaoran Zhang, Yijie Zhang, Jingxi Li, Xilin Yang, Wenjie Dong, Morgan Angus Darrow, Elham Kamangar, Han Sung Lee, Yair Rivenson, and Aydogan Ozcan, “Label-free virtual HER2 immunohistochemical staining of breast tissue using deep learning”, BME Frontiers (2022) https://spj.sciencemag.org/journals/bmef/2022/9786242/
