Examples of Past Undergraduate Research Projects
Wide-field Computational Imaging of Pathology Slides using Lensfree On-Chip Microscopy

Students: A. Feizi, P. Chung Advisor: Aydogan Ozcan Project Timeframe: 2014
Optical examination of microscale features in pathology slides is one of the gold standards to diagnose disease. However, the use of conventional light microscopes is partially limited owing to their relatively high cost, bulkiness of lens-based optics, small field of view (FOV), and requirements for lateral scanning and three-dimensional (3D) focus adjustment. We illustrate the performance of a computational lens-free, holographic on-chip microscope that uses the transport-of-intensity equation, multi-height iterative phase retrieval, and rotational field transformations to perform wide-FOV imaging of pathology samples with comparable image quality to a traditional transmission lens-based microscope. Using this lens-free on-chip microscope, we successfully imaged invasive carcinoma cells within human breast sections, Papanicolaou smears revealing a high-grade squamous intraepithelial lesion, and sickle cell anemia blood smears over a FOV of 20.5 mm2. By providing high-resolution images of large-area pathology samples with 3D digital focus adjustment, lens-free on-chip microscopy can be useful in resource-limited and point-of-care settings.
Imaging and Sizing of Single DNA Molecules on a Mobile-Phone

Students: S. Chiang, T. Kappel, C. Mejia and E. Yan Advisor: Aydogan Ozcan Project Timeframe: 2014
We demonstrate imaging and length quantification of single molecule DNA strands using a compact, lightweight and cost-effective fluorescence microscope installed on a mobile phone. In addition to an optomechanical attachment that creates a high contrast dark-field imaging setup using an external lens, thin-film interference filters, a miniature dovetail stage and a laser-diode for oblique-angle excitation, we also created a computational framework and a mobile phone application connected to a server back-end for measurement of the lengths of individual DNA molecules that are labeled and stretched using disposable chips. Using this mobile phone platform, we imaged single DNA molecules of various lengths to demonstrate a sizing accuracy of <1 kilobase-pairs (kbp) for 10 kbp and longer DNA samples imaged over a field-of-view of ∼2 mm2.
Design and modeling of graphene composite right/left handed metamaterial waveguides
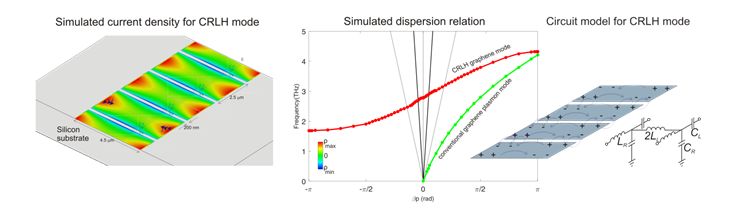
Student: Derrick Chu Advisor: Benjamin Williams Project Timeframe: 2014-2015
The goal of this project was to design and simulate composite right-left handed (CRLH) metamaterial waveguides based upon graphene plasmons, and evaluate their feasibility for waveguiding, phase shifting, and leaky-wave antennas in the THz and mid-infrared range. CRLH metamaterials can exhibit unique travelling wave behavior, including left-handed (i.e. negative refractive index), zero-index, and right-handed (i.e. positive refractive index) propagation, depending upon the frequency of excitation.
Journal Publication: Derrick A. Chu, P. W. C. Hon, T. Itoh, and B. S. Williams, “Feasibility of graphene CRLH metamaterial waveguides and leaky wave antennas,” Journal of Applied Physics vol 120, 013103 (2016).
Estimation of Subglottal Resonances for Children’s Speech Analysis
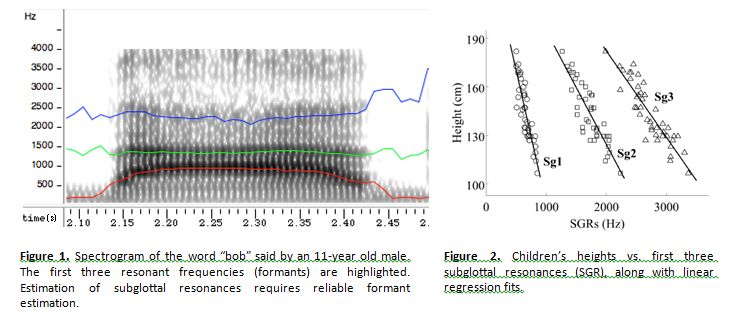
Student: Gary Yeung Advisor: Abeer Alwan Project Timeframe: 2015
The goal of this project was to produce algorithms to approximate the subglottal resonances of children using only the speech signal and apply these estimation algorithms to various applications. Applications of subglottal resonances include speaker normalization for speech recognition, speaker recognition, height estimation, and age estimation. Work supported in part by NSF Grant No. 0905381.
Club-Foot Physical Therapy Active Monitor
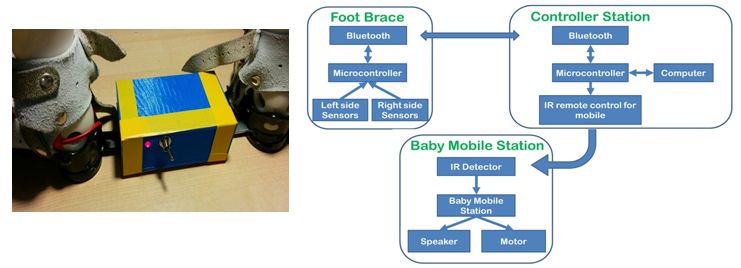
Students: Nicolò Maganzini, Geronimo Filippini, Alexandria Petterson Advisor: Greg Pottie Project Timeframe: 2014-2015
Newborns affected by clubfoot often undergo surgery which is followed by a prolonged period in which the patient must wear a foot brace. More rapid and effective recovery could be achieved by training newborns to exercise their leg muscles. This device monitors and detects correct movements of the patient’s foot within the brace and initiates a reward mechanism to teach the newborn to repeat it. Pressure sensors placed within the brace are used to detect foot movements. A μ-controller on the brace collects data and transfers it over Bluetooth to another controller. When the correct movement is detected, a moving baby-mobile mounted on the crib is activated, rewarding the infant. A USB connection and software allows for data collection, sensor calibration, and pressure threshold adjustment to accommodate for stronger movements as the training progresses. Clinical testing will begin soon with patients from the UCLA Orthopaedic Hospital.
Publication: Science poster day presentation and demonstration. Dean’s Prize for best demonstration.
Algorithms for unconditional security over networks
Student: Shengkang Chen Advisor: Martina Cardone, Christina Fragouli Project Timeframe: 2015-2016
Nowadays, we increasingly rely on wireless for services that are crucial to be private and secure (health, banking, professional). At the same time, it is of critical importance to explore methods for security that do not rely on the computational limitations of the adversaries. This project is part of a research direction that explores algorithms and methods for ensuring information theoretical (unconditional) security over networks. During this project we compared the performance of algorithms that we have designed, and showed that we can always do as well as – and sometimes many times much better than – the best currently available schemes in terms of secrecy rate.
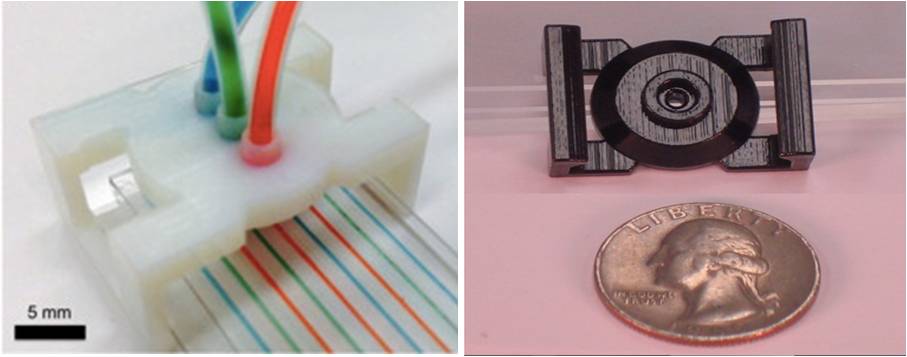
3D-printing for microfluidic channels and interconnects
Students: Carlo Paredes, Neal Shah Advisor: Rob Candler Project Timeframe: 2011-2014
Major issues in healthcare today deal with cost and accessibility of care to locations with limited resources. In order to address this growing concern, we investigated methods to interface with lab-on-chip devices—which allow lab tests to be conducted on miniature devices with small amounts of sample and expensive reagents— by using 3D printing to create fluidic connections to microfluidic chips. Utilization of this method allows the user to insert a minimal amount of each specimen into a lab-on-chip, and still obtain accurate results. Our microfluidic interconnection allows communication between the macro scale and the micro scale through creating a high-pressure fluid-tight seal. These qualities ensure uses in real world diagnostic applications, such as miniaturized medical laboratories.
Publications:
Carlo and Neal were invited to attend the “Posters On the Hill” event with Prof. Candler, where they presented their research to members of the U.S. Congress and their staff.
O. H. Paydar, C. N. Paredes, R. N. Candler, “Three Dimensionally-Printed (3DP) Microfluidic Devices”, Proceedings of Solid State Sensors, Actuators, and Microsystems Workshop (Hilton Head), 2012.
C. N. Paredes, J. Paz, N. Shah, R. N. Candler, “Microfluidic Chip Interconnect for Research and Diagnostics,” presented at LabAutomation2011, 2011.
Visualization Tools for Data Synchronization
Students: Chi-Yo Tsai Advisor: Lara Dolecek Project Timeframe: 2015-2016
Large-scale data is known to contain many redundancies although identifying redundancies and overlaps among different files is a difficult problem, due to file edits. We have developed an algorithm for efficient file synchronization under edit errors. The undergraduate research aspect of the this project focused on the development of visualization tools that illustrate the main algorithm – these tools are particularly useful for approximate synchronization of text and image files.