Virtual staining of unlabeled autopsy tissue using AI
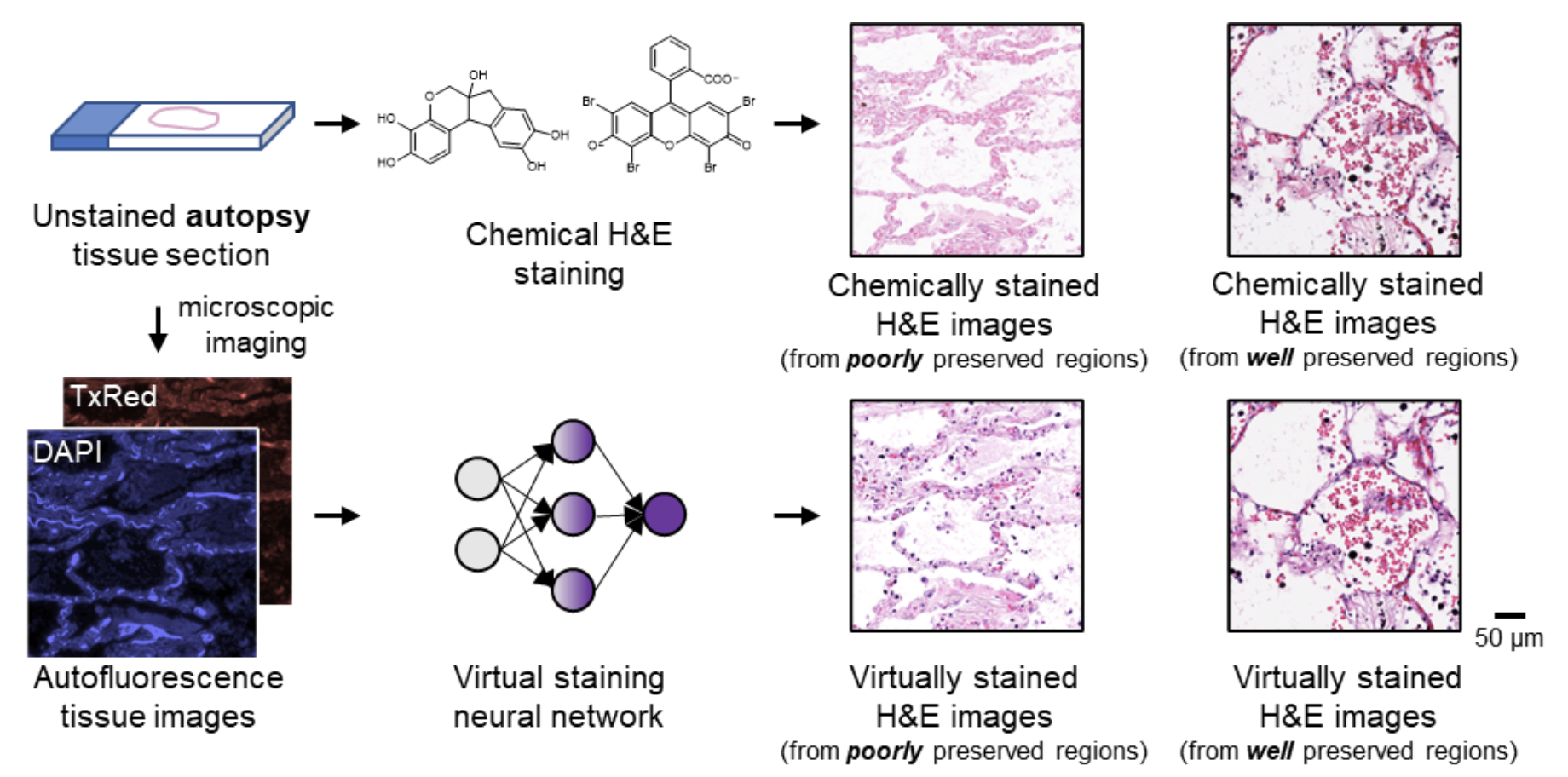
Autopsy has a central role in shedding light on diseases, helping to uncover the cause of death. Tissue samples from various organs are sampled, stained, and examined under a light microscope to evaluate their histological characteristics. However, chemical staining of autopsy samples often suffers from fixation-related artifacts due to unavoidable delays in tissue preservation. Tissue fixation delay leads to autolysis, a cellular self-digestion process, resulting in a series of perturbations in the tissue environment. These changes interfere with the normal binding of staining dyes and biomolecules in the tissue, thereby diminishing the quality of chemical staining and impacting the clarity and reliability of autopsy outcomes. Moreover, the current workflow for chemical staining is costly, time-consuming, and labor-intensive. Meeting the quality control demands of staining reagents, laboratory infrastructure, and skilled labor becomes challenging during global health emergencies, such as the COVID-19 pandemic, when histopathology services become limited, exacerbating the delays in post-mortem processing. These factors increase staining artifacts observed in autopsy samples.
In a recent work published in Nature Communications, a UCLA research team led by Professor Aydogan Ozcan developed an AI-powered computational staining approach for label-free autopsy tissue samples, which digitally transforms autofluorescence microscopic images of autopsy tissue sections into their high-quality hematoxylin and eosin (H&E) stained counterparts using a trained neural network model. This virtual staining method was proven to mitigate autolysis-induced artifacts inherent in traditional chemical staining of autopsy samples. It revealed the nuclear, cytoplasmic, and extracellular features clearly, particularly in tissue samples severely affected by autolysis, such as COVID-19 samples, where notable delays in tissue preservation occurred.
To further demonstrate the effectiveness of this autopsy virtual staining technique and ensure its superior performance, four board-certified pathologists blindly evaluated the staining quality of test images from autopsy samples without knowing which image was virtually stained or chemically stained. They confirmed that there was no significant difference between the virtually stained and chemically stained H&E images for the well-preserved autopsy tissue regions in terms of nuclear detail, cytoplasmic detail, and extracellular detail, underscoring the efficacy of this technique. In addition to mitigating autolysis-induced artifacts observed in traditional chemical staining of autopsy samples, this virtual staining technique is also faster, more repeatable and cost-effective, eliminating toxic reagents and manual labor involved in the traditional staining workflow, and therefore holds substantial potential for widespread applications in histology field.
See the article:
Yuzhu Li, Nir Pillar, Jingxi Li, Tairan Liu, Di Wu, Songyu Sun, Guangdong Ma, Kevin de Haan, Luzhe Huang, Yijie Zhang, Sepehr Hamidi, Anatoly Urisman, Tal Keidar Haran, William Dean Wallace, Jonathan E. Zuckerman, and Aydogan Ozcan, “Virtual histological staining of unlabeled autopsy tissue”, Nature Communications (2024)
https://www.nature.com/articles/s41467-024-46077-2
